import pandas as pdNB: Introducting Pandas
What is Pandas?
Pandas is a Python library design to work with dataframes.
Essentially, it adds a ton of usability features to NumPy.
It has become a standard library in data science.
Pandas Data Frames
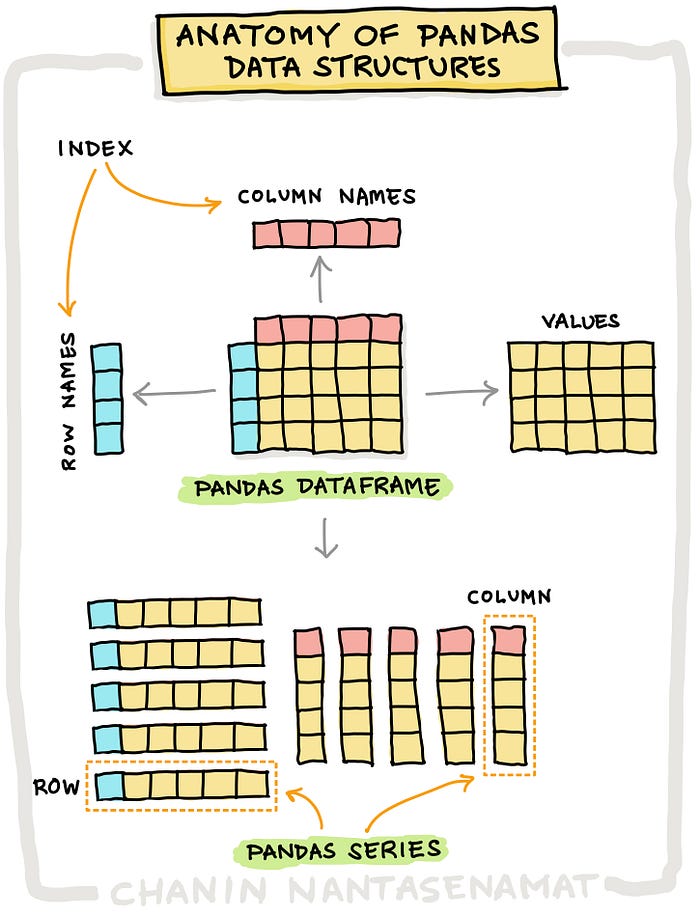
Just as NumPy introduces the n-dimensional array as a new data structure to Python, so Pandas introduces two:
The Series: a 1-dimensional labeled array capable of holding any data type.
The DataFrame: a 2-dimensional labeled array with columns of potentially different types.
By far, the most important data structure in Pandas is the dataframe (sometimes spelled “data frame”), with the series playing a supporting – but crucial – role.
In fact, dataframe objects are built out of series objects.
So, to understand what a dataframe is and how it behaves, you need to understand what is series is and how it is constructed.
Before going into that, here are two quick observations about dataframes:
First, dataframes are inspired by the R structure of the same name.
They have many similarities, but there are fundamental differences between the two that go beyond mere language differences.
Most important is the Pandas dataframes have indexes, whereas R dataframes do not.
Second, it is helpful to think of Pandas as wrapper around NumPy and Matplotlib that makes it much easier to perform common operations, like select data by column name or visualizing plots.
But this comes at a cost – Pandas is slower than NumPy.
This represents the classic trade-off between ease-of-use for humnas and machine performance.
Series Objects
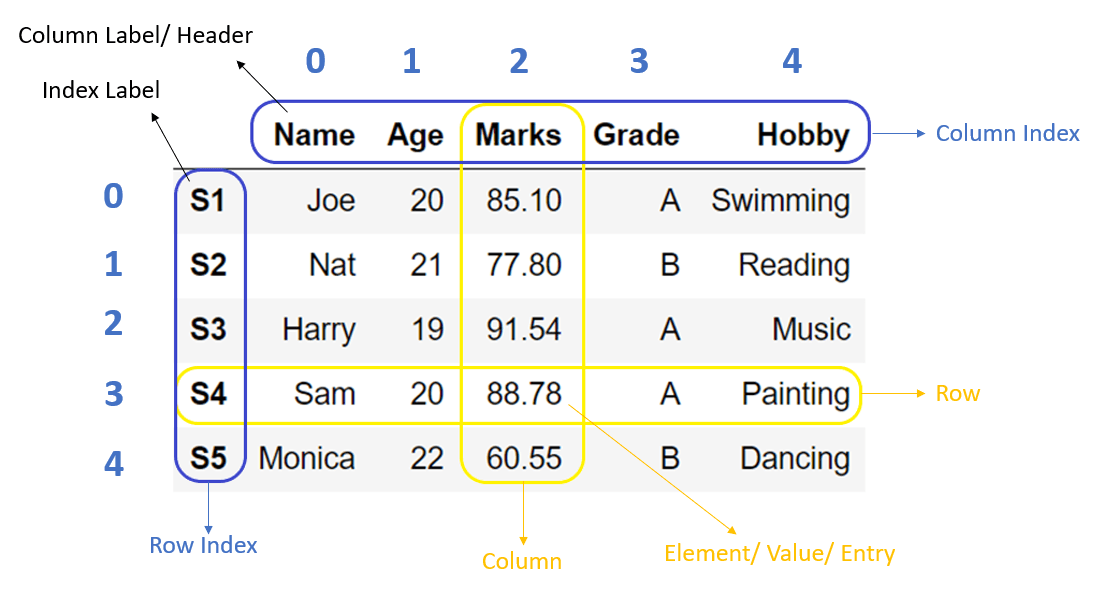
Axis Labels (Indexes)
A series is at heart a one-dimensional array with labels along its axis.
- Labels are essentially names that, ideally, uniquely identify each row (observation).
- It’s data must be of a single type, like NumPy arrays (which they are internally).
- The axis labels are collectively referred to as the index.
Think of the index as a separate data structure that is attached to the array. * The array holds the data. * The index holds the names of the observations or things that the data are about.
Why have an index?
- Indexes provide a way to access elements of the array by name.
- They allows series objects that share index labels to be combined.
- Many other things …
In fact, a dataframe is a collection of series with a common index.
To this collection of series, the dataframe also adds a set of labels along the horizontal axis. * The row index is axis 0. * The column index is called axis 1.
Note that both index and column labels can be multidimensional. * The are called Hierarchical Indexes and go the technical name of MultiIndexes. * As an example, consider that a table of text data might have a two-column index: (book_id, chap_id) * See the Pandas documentation.
It is crucial to understand the difference between the index of a dataframe and its data in order to understand how dataframes work.
Many a headache is caused by not understanding this difference :-)
Indexes are powerful and controversial. * They allow for all kinds of magic to take place when combining and accessing data. * But they are expensive and sometimes hard to work with (especially multiindexes). * They are especially difficult if you are coming from R and expecting dataframes to behave a certain way.
Some visuals to help
But enough introduction.
Let’s dive into how Pandas objects work in practice.
Importing
We import pandas like this, using the alias pd by convention:
We almost always import NumPy, too, since we use many of its functions with Pandas.
import numpy as npData Frame Constructors
There are several ways to create pandas data frames.
Passing a dictionary of lists:
df = pd.DataFrame({
'x': [0, 2, 1, 5],
'y': [1, 1, 0, 0],
'z': [True, False, False, False]
})df| x | y | z | |
|---|---|---|---|
| 0 | 0 | 1 | True |
| 1 | 2 | 1 | False |
| 2 | 1 | 0 | False |
| 3 | 5 | 0 | False |
df.indexRangeIndex(start=0, stop=4, step=1)list(df.index)[0, 1, 2, 3]df.columnsIndex(['x', 'y', 'z'], dtype='object')list(df.columns)['x', 'y', 'z']df.valuesarray([[0, 1, True],
[2, 1, False],
[1, 0, False],
[5, 0, False]], dtype=object)type(df.values)numpy.ndarrayPassing the three required pieces: - columns as list - index as list - data as list of lists (2D)
df2 = pd.DataFrame(
columns=['x','y'],
index=['row1','row2','row3'],
data=[[9,3],[1,2],[4,6]])df2| x | y | |
|---|---|---|
| row1 | 9 | 3 |
| row2 | 1 | 2 |
| row3 | 4 | 6 |
Passing a list of tuples (or list-like objects):
my_data = [
('a', 1, True),
('b', 2, False)
]
df3 = pd.DataFrame(my_data, columns=['f1', 'f2', 'f3'])df3| f1 | f2 | f3 | |
|---|---|---|---|
| 0 | a | 1 | True |
| 1 | b | 2 | False |
Naming indexes
It is helpful to name your indexes.
df3.index.name = 'obs_id'df3| f1 | f2 | f3 | |
|---|---|---|---|
| obs_id | |||
| 0 | a | 1 | True |
| 1 | b | 2 | False |
Copying DataFrames with copy()
Use copy() to give the new df a clean break from the original.
Otherwise, the copied df will point to the same object as the original.
df = pd.DataFrame(
{
'x':[0,2,1,5],
'y':[1,1,0,0],
'z':[True,False,False,False]
}
) df| x | y | z | |
|---|---|---|---|
| 0 | 0 | 1 | True |
| 1 | 2 | 1 | False |
| 2 | 1 | 0 | False |
| 3 | 5 | 0 | False |
We create two copies, one “deep” and one “shallow”.
df_deep = df.copy() # deep copy; changes to df will not pass through
df_shallow = df # shallow copy; changes to df will pass throughIf we alter a value in the original …
df.x = 1df| x | y | z | |
|---|---|---|---|
| 0 | 1 | 1 | True |
| 1 | 1 | 1 | False |
| 2 | 1 | 0 | False |
| 3 | 1 | 0 | False |
… then the shallow copy is also changed …
df_shallow| x | y | z | |
|---|---|---|---|
| 0 | 1 | 1 | True |
| 1 | 1 | 1 | False |
| 2 | 1 | 0 | False |
| 3 | 1 | 0 | False |
… while the deep copy is not.
df_deep| x | y | z | |
|---|---|---|---|
| 0 | 0 | 1 | True |
| 1 | 2 | 1 | False |
| 2 | 1 | 0 | False |
| 3 | 5 | 0 | False |
Of course, the reverse is true too – changes to the shallow copy affect the original:
df_shallow.y = 99df| x | y | z | |
|---|---|---|---|
| 0 | 1 | 99 | True |
| 1 | 1 | 99 | False |
| 2 | 1 | 99 | False |
| 3 | 1 | 99 | False |
So, df_shallow mirrors changes to df, since it references its indices and data.
df_deep does not reference df, and so changes to df do not impact df_deep.
Let’s reset our dataframe.
df = pd.DataFrame({'x':[0,2,1,5], 'y':[1,1,0,0], 'z':[True,False,False,False]}) Column Data Types
With .types
df.dtypesx int64
y int64
z bool
dtype: objectWith .info()
df.info()<class 'pandas.core.frame.DataFrame'>
RangeIndex: 4 entries, 0 to 3
Data columns (total 3 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 x 4 non-null int64
1 y 4 non-null int64
2 z 4 non-null bool
dtypes: bool(1), int64(2)
memory usage: 200.0 bytesColumn Renaming
Can rename one or more fields at once using a dict.
Rename the field z to is_label:
df = df.rename(columns={'z': 'is_label'})df| x | y | is_label | |
|---|---|---|---|
| 0 | 0 | 1 | True |
| 1 | 2 | 1 | False |
| 2 | 1 | 0 | False |
| 3 | 5 | 0 | False |
You can also change column names this way:
old_cols = df.columns # Keep a copy so we can revert
df.columns = ['X','Y', 'LABEL']df| X | Y | LABEL | |
|---|---|---|---|
| 0 | 0 | 1 | True |
| 1 | 2 | 1 | False |
| 2 | 1 | 0 | False |
| 3 | 5 | 0 | False |
df.columns = old_cols # Reset thingsColumn Referencing
Pandas supports both bracket notation and dot notation.
Bracket
df['y']0 1
1 1
2 0
3 0
Name: y, dtype: int64Dot (i.e. as object attribute)
df.y0 1
1 1
2 0
3 0
Name: y, dtype: int64Dot notation is very convenient, since as object attributes they can be tab-completed in various editing environments.
But: - It only works if the column names are not reserved words. - It can’t be used when creating a new column (see below).
It is convenient to names columns with a prefix, e.g. doc_title, doc_year, doc_author, etc. to avoid name collisions.
Column attributes and methods work with both:
df.y.values, df['y'].values(array([1, 1, 0, 0]), array([1, 1, 0, 0]))show only the first value, by indexing:
df.y.values[0]1Column Selection
You select columns from a dataframe by passing a value or list (or any expression that evaluates to a list).
Calling a columns with a scalar returns a Series:
df['x']0 0
1 2
2 1
3 5
Name: x, dtype: int64type(df['x'])pandas.core.series.SeriesCalling a column with a list returns a dataframe:
df[['x']]| x | |
|---|---|
| 0 | 0 |
| 1 | 2 |
| 2 | 1 |
| 3 | 5 |
type(df[['x']])pandas.core.frame.DataFrameIn Pandas, we can use “fancy indexing” with labels:
df[['y', 'x']]| y | x | |
|---|---|---|
| 0 | 1 | 0 |
| 1 | 1 | 2 |
| 2 | 0 | 1 |
| 3 | 0 | 5 |
We can put in a list comprehension, too:
df[[col for col in df.columns if col not in ['x','y']]]| is_label | |
|---|---|
| 0 | True |
| 1 | False |
| 2 | False |
| 3 | False |
Adding New Columns
It is typical to create a new column from existing columns.
In this example, a new column (or field) is created by summing x and y:
df['x_plus_y'] = df.x + df.ydf| x | y | is_label | x_plus_y | |
|---|---|---|---|---|
| 0 | 0 | 1 | True | 1 |
| 1 | 2 | 1 | False | 3 |
| 2 | 1 | 0 | False | 1 |
| 3 | 5 | 0 | False | 5 |
Note the use of bracket notation on the left.
When new columns are created, you must use bracket notation.
Removing Columns with del and .drop()
del
del can drop a DataFrame or single columns from the frame
df_drop = df.copy()df_drop.head(2)| x | y | is_label | x_plus_y | |
|---|---|---|---|---|
| 0 | 0 | 1 | True | 1 |
| 1 | 2 | 1 | False | 3 |
del df_drop['x']df_drop| y | is_label | x_plus_y | |
|---|---|---|---|
| 0 | 1 | True | 1 |
| 1 | 1 | False | 3 |
| 2 | 0 | False | 1 |
| 3 | 0 | False | 5 |
.drop()
Can drop one or more columns.
takes axis parameter: - axis=0 refers to rows
- axis=1 refers to columns
df_drop = df_drop.drop(['x_plus_y', 'is_label'], axis=1)df_drop| y | |
|---|---|
| 0 | 1 |
| 1 | 1 |
| 2 | 0 |
| 3 | 0 |
Load Iris Dataset
Let’s load a bigger data set to explore more functionality.
The function load_dataset() in the seaborn package loads the built-in dataset.
import seaborn as sns
iris = sns.load_dataset('iris')Check the data type of iris:
type(iris)pandas.core.frame.DataFrameSee the first and last records with .head() and .tail()
iris.head()| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
iris.head(10)| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
| 5 | 5.4 | 3.9 | 1.7 | 0.4 | setosa |
| 6 | 4.6 | 3.4 | 1.4 | 0.3 | setosa |
| 7 | 5.0 | 3.4 | 1.5 | 0.2 | setosa |
| 8 | 4.4 | 2.9 | 1.4 | 0.2 | setosa |
| 9 | 4.9 | 3.1 | 1.5 | 0.1 | setosa |
iris.tail()| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| 145 | 6.7 | 3.0 | 5.2 | 2.3 | virginica |
| 146 | 6.3 | 2.5 | 5.0 | 1.9 | virginica |
| 147 | 6.5 | 3.0 | 5.2 | 2.0 | virginica |
| 148 | 6.2 | 3.4 | 5.4 | 2.3 | virginica |
| 149 | 5.9 | 3.0 | 5.1 | 1.8 | virginica |
Inspect metadata
iris.dtypessepal_length float64
sepal_width float64
petal_length float64
petal_width float64
species object
dtype: objectshape (rows, columns):
iris.shape(150, 5)Alternatively, len() returns row (record) count:
len(iris)150Column names:
iris.columnsIndex(['sepal_length', 'sepal_width', 'petal_length', 'petal_width',
'species'],
dtype='object')Get it all with .info()
iris.info()<class 'pandas.core.frame.DataFrame'>
RangeIndex: 150 entries, 0 to 149
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 sepal_length 150 non-null float64
1 sepal_width 150 non-null float64
2 petal_length 150 non-null float64
3 petal_width 150 non-null float64
4 species 150 non-null object
dtypes: float64(4), object(1)
memory usage: 6.0+ KBThe Index
iris.indexRangeIndex(start=0, stop=150, step=1)We can name indexes, and it is important to do so in many cases.
iris.index.name = 'obs_id' # Each observation is a unique plantiris| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| obs_id | |||||
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
| ... | ... | ... | ... | ... | ... |
| 145 | 6.7 | 3.0 | 5.2 | 2.3 | virginica |
| 146 | 6.3 | 2.5 | 5.0 | 1.9 | virginica |
| 147 | 6.5 | 3.0 | 5.2 | 2.0 | virginica |
| 148 | 6.2 | 3.4 | 5.4 | 2.3 | virginica |
| 149 | 5.9 | 3.0 | 5.1 | 1.8 | virginica |
150 rows × 5 columns
We can also redefine indexes to reflect the logic of our data.
In this data set, the species of the flower is part of its identity, so it can be part of the index.
The other features vary by individual.
Note that species is also a label that can be used for training a model to predict the species of an iris flower. In that use case, the column would be pulled out into a separate vector.
iris_w_idx = iris.reset_index().set_index(['species','obs_id'])iris_w_idx| sepal_length | sepal_width | petal_length | petal_width | ||
|---|---|---|---|---|---|
| species | obs_id | ||||
| setosa | 0 | 5.1 | 3.5 | 1.4 | 0.2 |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | |
| ... | ... | ... | ... | ... | ... |
| virginica | 145 | 6.7 | 3.0 | 5.2 | 2.3 |
| 146 | 6.3 | 2.5 | 5.0 | 1.9 | |
| 147 | 6.5 | 3.0 | 5.2 | 2.0 | |
| 148 | 6.2 | 3.4 | 5.4 | 2.3 | |
| 149 | 5.9 | 3.0 | 5.1 | 1.8 |
150 rows × 4 columns
Row Selection (Filtering)
iloc[]
You can extract rows using indexes with iloc[].
This fetches row 3, and all columns.
iris.iloc[2]sepal_length 4.7
sepal_width 3.2
petal_length 1.3
petal_width 0.2
species setosa
Name: 2, dtype: objectfetch rows with indices 1,2 (the right endpoint is exclusive), and all columns.
iris.iloc[1:3]| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| obs_id | |||||
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
fetch rows with indices 1,2 and first three columns (positions 0, 1, 2)
Combining Filtering and Selecting
So, remember the comma notation from NumPy – it is used here.
The first element is a row selector, the second a column selector.
In database terminology, row selection is called filtering.
iris.iloc[1:3, 0:3]| sepal_length | sepal_width | petal_length | |
|---|---|---|---|
| obs_id | |||
| 1 | 4.9 | 3.0 | 1.4 |
| 2 | 4.7 | 3.2 | 1.3 |
You can apply slices to column names too. You don’t need .iloc[] here.
iris.columns[0:3]Index(['sepal_length', 'sepal_width', 'petal_length'], dtype='object').loc[]
Filtering can also be done with .loc[]. This uses the row, column labels (names).
Here we ask for rows with labels (indexes) 1-3, and it gives exactly that
.iloc[] returned rows with indices 1,2.
Author note: This is by far the more useful of the two in my experience.
iris.loc[1:3]| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| obs_id | |||||
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
Note the different behavior of the slice here – with .loc, 1:3 is short-hand for [1,2,3].
iris.loc[[1,2,3]]| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| obs_id | |||||
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
So, we are not using normal slicing here:
iris.loc[[:-1]]SyntaxError: invalid syntax (170941475.py, line 1)Although this works:
iris.loc[:]| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| obs_id | |||||
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
| ... | ... | ... | ... | ... | ... |
| 145 | 6.7 | 3.0 | 5.2 | 2.3 | virginica |
| 146 | 6.3 | 2.5 | 5.0 | 1.9 | virginica |
| 147 | 6.5 | 3.0 | 5.2 | 2.0 | virginica |
| 148 | 6.2 | 3.4 | 5.4 | 2.3 | virginica |
| 149 | 5.9 | 3.0 | 5.1 | 1.8 | virginica |
150 rows × 5 columns
Take-away: Fancy indexing uses new lists; you can use something like [:-1] because you are not referring to an existing list.
Subset on columns with column name (as a string) or list of strings
iris.loc[1:3, ['sepal_length','petal_width']]| sepal_length | petal_width | |
|---|---|---|
| obs_id | ||
| 1 | 4.9 | 0.2 |
| 2 | 4.7 | 0.2 |
| 3 | 4.6 | 0.2 |
Select all rows, specific columns
iris.loc[:, ['sepal_length','petal_width']]| sepal_length | petal_width | |
|---|---|---|
| obs_id | ||
| 0 | 5.1 | 0.2 |
| 1 | 4.9 | 0.2 |
| 2 | 4.7 | 0.2 |
| 3 | 4.6 | 0.2 |
| 4 | 5.0 | 0.2 |
| ... | ... | ... |
| 145 | 6.7 | 2.3 |
| 146 | 6.3 | 1.9 |
| 147 | 6.5 | 2.0 |
| 148 | 6.2 | 2.3 |
| 149 | 5.9 | 1.8 |
150 rows × 2 columns
.loc[] with MultiIndex
Recall our dataframe with a two element index:
iris_w_idx| sepal_length | sepal_width | petal_length | petal_width | ||
|---|---|---|---|---|---|
| species | obs_id | ||||
| setosa | 0 | 5.1 | 3.5 | 1.4 | 0.2 |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | |
| ... | ... | ... | ... | ... | ... |
| virginica | 145 | 6.7 | 3.0 | 5.2 | 2.3 |
| 146 | 6.3 | 2.5 | 5.0 | 1.9 | |
| 147 | 6.5 | 3.0 | 5.2 | 2.0 | |
| 148 | 6.2 | 3.4 | 5.4 | 2.3 | |
| 149 | 5.9 | 3.0 | 5.1 | 1.8 |
150 rows × 4 columns
Selecting a single observation by it’s key, i.e. full label:
iris_w_idx.loc[('setosa',0)] # df.at[r,c]sepal_length 5.1
sepal_width 3.5
petal_length 1.4
petal_width 0.2
Name: (setosa, 0), dtype: float64Selecting just the setosas:
iris_w_idx.loc['setosa']| sepal_length | sepal_width | petal_length | petal_width | |
|---|---|---|---|---|
| obs_id | ||||
| 0 | 5.1 | 3.5 | 1.4 | 0.2 |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 |
| 5 | 5.4 | 3.9 | 1.7 | 0.4 |
| 6 | 4.6 | 3.4 | 1.4 | 0.3 |
| 7 | 5.0 | 3.4 | 1.5 | 0.2 |
| 8 | 4.4 | 2.9 | 1.4 | 0.2 |
| 9 | 4.9 | 3.1 | 1.5 | 0.1 |
| 10 | 5.4 | 3.7 | 1.5 | 0.2 |
| 11 | 4.8 | 3.4 | 1.6 | 0.2 |
| 12 | 4.8 | 3.0 | 1.4 | 0.1 |
| 13 | 4.3 | 3.0 | 1.1 | 0.1 |
| 14 | 5.8 | 4.0 | 1.2 | 0.2 |
| 15 | 5.7 | 4.4 | 1.5 | 0.4 |
| 16 | 5.4 | 3.9 | 1.3 | 0.4 |
| 17 | 5.1 | 3.5 | 1.4 | 0.3 |
| 18 | 5.7 | 3.8 | 1.7 | 0.3 |
| 19 | 5.1 | 3.8 | 1.5 | 0.3 |
| 20 | 5.4 | 3.4 | 1.7 | 0.2 |
| 21 | 5.1 | 3.7 | 1.5 | 0.4 |
| 22 | 4.6 | 3.6 | 1.0 | 0.2 |
| 23 | 5.1 | 3.3 | 1.7 | 0.5 |
| 24 | 4.8 | 3.4 | 1.9 | 0.2 |
| 25 | 5.0 | 3.0 | 1.6 | 0.2 |
| 26 | 5.0 | 3.4 | 1.6 | 0.4 |
| 27 | 5.2 | 3.5 | 1.5 | 0.2 |
| 28 | 5.2 | 3.4 | 1.4 | 0.2 |
| 29 | 4.7 | 3.2 | 1.6 | 0.2 |
| 30 | 4.8 | 3.1 | 1.6 | 0.2 |
| 31 | 5.4 | 3.4 | 1.5 | 0.4 |
| 32 | 5.2 | 4.1 | 1.5 | 0.1 |
| 33 | 5.5 | 4.2 | 1.4 | 0.2 |
| 34 | 4.9 | 3.1 | 1.5 | 0.2 |
| 35 | 5.0 | 3.2 | 1.2 | 0.2 |
| 36 | 5.5 | 3.5 | 1.3 | 0.2 |
| 37 | 4.9 | 3.6 | 1.4 | 0.1 |
| 38 | 4.4 | 3.0 | 1.3 | 0.2 |
| 39 | 5.1 | 3.4 | 1.5 | 0.2 |
| 40 | 5.0 | 3.5 | 1.3 | 0.3 |
| 41 | 4.5 | 2.3 | 1.3 | 0.3 |
| 42 | 4.4 | 3.2 | 1.3 | 0.2 |
| 43 | 5.0 | 3.5 | 1.6 | 0.6 |
| 44 | 5.1 | 3.8 | 1.9 | 0.4 |
| 45 | 4.8 | 3.0 | 1.4 | 0.3 |
| 46 | 5.1 | 3.8 | 1.6 | 0.2 |
| 47 | 4.6 | 3.2 | 1.4 | 0.2 |
| 48 | 5.3 | 3.7 | 1.5 | 0.2 |
| 49 | 5.0 | 3.3 | 1.4 | 0.2 |
Grabbing one species and one feature:
iris_w_idx.loc['setosa', 'sepal_length'].head()obs_id
0 5.1
1 4.9
2 4.7
3 4.6
4 5.0
Name: sepal_length, dtype: float64This returns a series. If we want a dataframe back, we can use .to_frame():
iris_w_idx.loc['setosa', 'sepal_length'].to_frame().head()| sepal_length | |
|---|---|
| obs_id | |
| 0 | 5.1 |
| 1 | 4.9 |
| 2 | 4.7 |
| 3 | 4.6 |
| 4 | 5.0 |
We use a tuple to index multiple index levels.
Note that you can’t pass slices here – and this where indexing can get sticky.
iris_w_idx.loc[('setosa', 5)]sepal_length 5.4
sepal_width 3.9
petal_length 1.7
petal_width 0.4
Name: (setosa, 5), dtype: float64Another Example
df_cat = pd.DataFrame(
index=['burmese', 'persian', 'maine_coone'],
columns=['x'],
data=[2,1,3]
)df_cat| x | |
|---|---|
| burmese | 2 |
| persian | 1 |
| maine_coone | 3 |
df_cat.iloc[:2]| x | |
|---|---|
| burmese | 2 |
| persian | 1 |
df_cat.iloc[0:1]| x | |
|---|---|
| burmese | 2 |
df_cat.loc['burmese']x 2
Name: burmese, dtype: int64df_cat.loc[['burmese','maine_coone']]| x | |
|---|---|
| burmese | 2 |
| maine_coone | 3 |
Boolean Filtering
It’s very common to subset a dataframe based on some condition on the data.
Note that even though we are filtering rows, we are not using .loc[] or .iloc[] here.
Pandas knows what to do if you pass a boolean structure.
iris.sepal_length >= 7.5obs_id
0 False
1 False
2 False
3 False
4 False
...
145 False
146 False
147 False
148 False
149 False
Name: sepal_length, Length: 150, dtype: booliris[iris.sepal_length >= 7.5]| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| obs_id | |||||
| 105 | 7.6 | 3.0 | 6.6 | 2.1 | virginica |
| 117 | 7.7 | 3.8 | 6.7 | 2.2 | virginica |
| 118 | 7.7 | 2.6 | 6.9 | 2.3 | virginica |
| 122 | 7.7 | 2.8 | 6.7 | 2.0 | virginica |
| 131 | 7.9 | 3.8 | 6.4 | 2.0 | virginica |
| 135 | 7.7 | 3.0 | 6.1 | 2.3 | virginica |
iris[(iris.sepal_length >= 4.5) & (iris.sepal_length <= 4.7)]| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| obs_id | |||||
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 6 | 4.6 | 3.4 | 1.4 | 0.3 | setosa |
| 22 | 4.6 | 3.6 | 1.0 | 0.2 | setosa |
| 29 | 4.7 | 3.2 | 1.6 | 0.2 | setosa |
| 41 | 4.5 | 2.3 | 1.3 | 0.3 | setosa |
| 47 | 4.6 | 3.2 | 1.4 | 0.2 | setosa |
Masking
Here’s an example of masking using boolean conditions passed to the dataframe selector:
Here are the values for the feature sepal length:
iris.sepal_length.valuesarray([5.1, 4.9, 4.7, 4.6, 5. , 5.4, 4.6, 5. , 4.4, 4.9, 5.4, 4.8, 4.8,
4.3, 5.8, 5.7, 5.4, 5.1, 5.7, 5.1, 5.4, 5.1, 4.6, 5.1, 4.8, 5. ,
5. , 5.2, 5.2, 4.7, 4.8, 5.4, 5.2, 5.5, 4.9, 5. , 5.5, 4.9, 4.4,
5.1, 5. , 4.5, 4.4, 5. , 5.1, 4.8, 5.1, 4.6, 5.3, 5. , 7. , 6.4,
6.9, 5.5, 6.5, 5.7, 6.3, 4.9, 6.6, 5.2, 5. , 5.9, 6. , 6.1, 5.6,
6.7, 5.6, 5.8, 6.2, 5.6, 5.9, 6.1, 6.3, 6.1, 6.4, 6.6, 6.8, 6.7,
6. , 5.7, 5.5, 5.5, 5.8, 6. , 5.4, 6. , 6.7, 6.3, 5.6, 5.5, 5.5,
6.1, 5.8, 5. , 5.6, 5.7, 5.7, 6.2, 5.1, 5.7, 6.3, 5.8, 7.1, 6.3,
6.5, 7.6, 4.9, 7.3, 6.7, 7.2, 6.5, 6.4, 6.8, 5.7, 5.8, 6.4, 6.5,
7.7, 7.7, 6. , 6.9, 5.6, 7.7, 6.3, 6.7, 7.2, 6.2, 6.1, 6.4, 7.2,
7.4, 7.9, 6.4, 6.3, 6.1, 7.7, 6.3, 6.4, 6. , 6.9, 6.7, 6.9, 5.8,
6.8, 6.7, 6.7, 6.3, 6.5, 6.2, 5.9])And here are the boolean values generated by applying a comparison operator to those values:
mask = iris.sepal_length >= 7.5mask.valuesarray([False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, False, False, False,
False, False, False, False, False, False, True, False, False,
False, False, False, False, False, False, False, False, False,
True, True, False, False, False, True, False, False, False,
False, False, False, False, False, True, False, False, False,
True, False, False, False, False, False, False, False, False,
False, False, False, False, False, False])mask.values.astype('int')array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 1,
0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0])The two sets of values have the same shape.
We can now overlay the logical values over the numeric ones and keep only what is True:
iris.sepal_length[mask].valuesarray([7.6, 7.7, 7.7, 7.7, 7.9, 7.7])Working with Missing Data
Pandas primarily uses the data type np.nan from NumPy to represent missing data.
df_miss = pd.DataFrame({
'x': [2, np.nan, 1],
'y': [np.nan, np.nan, 6]}
)These values appear as NaNs:
df_miss| x | y | |
|---|---|---|
| 0 | 2.0 | NaN |
| 1 | NaN | NaN |
| 2 | 1.0 | 6.0 |
.dropna()
This will drop all rows with missing data in any column.
df_drop_all = df_miss.dropna()
df_drop_all| x | y | |
|---|---|---|
| 2 | 1.0 | 6.0 |
The subset parameter takes a list of column names to specify which columns should have missing values.
df_drop_x = df_miss.dropna(subset=['x'])
df_drop_x| x | y | |
|---|---|---|
| 0 | 2.0 | NaN |
| 2 | 1.0 | 6.0 |
.fillna()
This will replace missing values with whatever you set it to, e.g. \(0\)s.
We can pass the results of an operation – for example to peform simple imputation, we can replace missing values in each column with the median value of the respective column:
df_filled = df_miss.fillna(df_miss.median())df_filled| x | y | |
|---|---|---|
| 0 | 2.0 | 6.0 |
| 1 | 1.5 | 6.0 |
| 2 | 1.0 | 6.0 |
Sorting
.sort_values()
Sort by values - by parameter takes string or list of strings - ascending takes True or False - inplace will save sorted values into the df
iris.sort_values(by=['sepal_length','petal_width'], ascending=False)| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| obs_id | |||||
| 131 | 7.9 | 3.8 | 6.4 | 2.0 | virginica |
| 118 | 7.7 | 2.6 | 6.9 | 2.3 | virginica |
| 135 | 7.7 | 3.0 | 6.1 | 2.3 | virginica |
| 117 | 7.7 | 3.8 | 6.7 | 2.2 | virginica |
| 122 | 7.7 | 2.8 | 6.7 | 2.0 | virginica |
| ... | ... | ... | ... | ... | ... |
| 41 | 4.5 | 2.3 | 1.3 | 0.3 | setosa |
| 8 | 4.4 | 2.9 | 1.4 | 0.2 | setosa |
| 38 | 4.4 | 3.0 | 1.3 | 0.2 | setosa |
| 42 | 4.4 | 3.2 | 1.3 | 0.2 | setosa |
| 13 | 4.3 | 3.0 | 1.1 | 0.1 | setosa |
150 rows × 5 columns
.sort_index()
Sort by index. Example sorts by descending index
iris.sort_index(axis=0, ascending=False)| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| obs_id | |||||
| 149 | 5.9 | 3.0 | 5.1 | 1.8 | virginica |
| 148 | 6.2 | 3.4 | 5.4 | 2.3 | virginica |
| 147 | 6.5 | 3.0 | 5.2 | 2.0 | virginica |
| 146 | 6.3 | 2.5 | 5.0 | 1.9 | virginica |
| 145 | 6.7 | 3.0 | 5.2 | 2.3 | virginica |
| ... | ... | ... | ... | ... | ... |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
150 rows × 5 columns
Statistics
describe()
iris.describe()| sepal_length | sepal_width | petal_length | petal_width | |
|---|---|---|---|---|
| count | 150.000000 | 150.000000 | 150.000000 | 150.000000 |
| mean | 5.843333 | 3.057333 | 3.758000 | 1.199333 |
| std | 0.828066 | 0.435866 | 1.765298 | 0.762238 |
| min | 4.300000 | 2.000000 | 1.000000 | 0.100000 |
| 25% | 5.100000 | 2.800000 | 1.600000 | 0.300000 |
| 50% | 5.800000 | 3.000000 | 4.350000 | 1.300000 |
| 75% | 6.400000 | 3.300000 | 5.100000 | 1.800000 |
| max | 7.900000 | 4.400000 | 6.900000 | 2.500000 |
iris.describe().T| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| sepal_length | 150.0 | 5.843333 | 0.828066 | 4.3 | 5.1 | 5.80 | 6.4 | 7.9 |
| sepal_width | 150.0 | 3.057333 | 0.435866 | 2.0 | 2.8 | 3.00 | 3.3 | 4.4 |
| petal_length | 150.0 | 3.758000 | 1.765298 | 1.0 | 1.6 | 4.35 | 5.1 | 6.9 |
| petal_width | 150.0 | 1.199333 | 0.762238 | 0.1 | 0.3 | 1.30 | 1.8 | 2.5 |
iris.species.describe()count 150
unique 3
top setosa
freq 50
Name: species, dtype: objectiris.sepal_length.describe()count 150.000000
mean 5.843333
std 0.828066
min 4.300000
25% 5.100000
50% 5.800000
75% 6.400000
max 7.900000
Name: sepal_length, dtype: float64value_counts()
This is a highly useful function for showing the frequency for each distinct value.
Parameters give the ability to sort by count or index, normalize, and more.
iris.species.value_counts()setosa 50
versicolor 50
virginica 50
Name: species, dtype: int64SPECIES = iris.species.value_counts().to_frame('n')SPECIES| n | |
|---|---|
| setosa | 50 |
| versicolor | 50 |
| virginica | 50 |
Show percentages instead of counts
iris.species.value_counts(normalize=True)setosa 0.333333
versicolor 0.333333
virginica 0.333333
Name: species, dtype: float64The methods returns a series that can be converted into a dataframe.
SEPAL_LENGTH = iris.sepal_length.value_counts().to_frame('n')SEPAL_LENGTH.head()| n | |
|---|---|
| 5.0 | 10 |
| 5.1 | 9 |
| 6.3 | 9 |
| 5.7 | 8 |
| 6.7 | 8 |
You can run .value_counts() on a column to get a kind of histogram:
SEPAL_LENGTH.sort_index().plot.bar(figsize=(8,4), rot=45);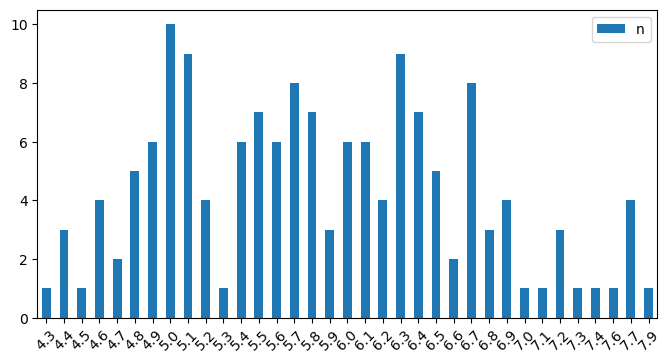
iris.sepal_length.hist();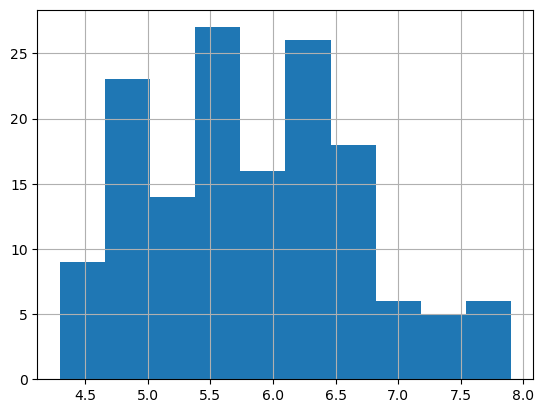
.mean()
Operations like this generally exclude missing data.
So, it is import to convert missing data to values if they need to be considered in the denominator.
iris.sepal_length.mean()5.843333333333334.max()
iris.sepal_length.max()7.9.std()
This standard deviation.
iris.sepal_length.std()0.8280661279778629.corr()
iris.corr()/var/folders/14/rnyfspnx2q131jp_752t9fc80000gn/T/ipykernel_1956/2141086772.py:1: FutureWarning: The default value of numeric_only in DataFrame.corr is deprecated. In a future version, it will default to False. Select only valid columns or specify the value of numeric_only to silence this warning.
iris.corr()| sepal_length | sepal_width | petal_length | petal_width | |
|---|---|---|---|---|
| sepal_length | 1.000000 | -0.117570 | 0.871754 | 0.817941 |
| sepal_width | -0.117570 | 1.000000 | -0.428440 | -0.366126 |
| petal_length | 0.871754 | -0.428440 | 1.000000 | 0.962865 |
| petal_width | 0.817941 | -0.366126 | 0.962865 | 1.000000 |
iris.corr(numeric_only=True)| sepal_length | sepal_width | petal_length | petal_width | |
|---|---|---|---|---|
| sepal_length | 1.000000 | -0.117570 | 0.871754 | 0.817941 |
| sepal_width | -0.117570 | 1.000000 | -0.428440 | -0.366126 |
| petal_length | 0.871754 | -0.428440 | 1.000000 | 0.962865 |
| petal_width | 0.817941 | -0.366126 | 0.962865 | 1.000000 |
Correlation can be computed on two fields by subsetting on them:
iris[['sepal_length','petal_length']].corr()| sepal_length | petal_length | |
|---|---|---|
| sepal_length | 1.000000 | 0.871754 |
| petal_length | 0.871754 | 1.000000 |
iris[['sepal_length','petal_length','sepal_width']].corr()| sepal_length | petal_length | sepal_width | |
|---|---|---|---|
| sepal_length | 1.000000 | 0.871754 | -0.11757 |
| petal_length | 0.871754 | 1.000000 | -0.42844 |
| sepal_width | -0.117570 | -0.428440 | 1.00000 |
Styling
iris.corr(numeric_only=True).style.background_gradient(cmap="Blues", axis=None)| sepal_length | sepal_width | petal_length | petal_width | |
|---|---|---|---|---|
| sepal_length | 1.000000 | -0.117570 | 0.871754 | 0.817941 |
| sepal_width | -0.117570 | 1.000000 | -0.428440 | -0.366126 |
| petal_length | 0.871754 | -0.428440 | 1.000000 | 0.962865 |
| petal_width | 0.817941 | -0.366126 | 0.962865 | 1.000000 |
iris.corr(numeric_only=True).style.bar(axis=None)| sepal_length | sepal_width | petal_length | petal_width | |
|---|---|---|---|---|
| sepal_length | 1.000000 | -0.117570 | 0.871754 | 0.817941 |
| sepal_width | -0.117570 | 1.000000 | -0.428440 | -0.366126 |
| petal_length | 0.871754 | -0.428440 | 1.000000 | 0.962865 |
| petal_width | 0.817941 | -0.366126 | 0.962865 | 1.000000 |
Visualization
Scatterplot using Seabprn on the df columns sepal_length, petal_length.
Visualization will be covered separately in more detail.
iris.plot.scatter('sepal_length', 'petal_length');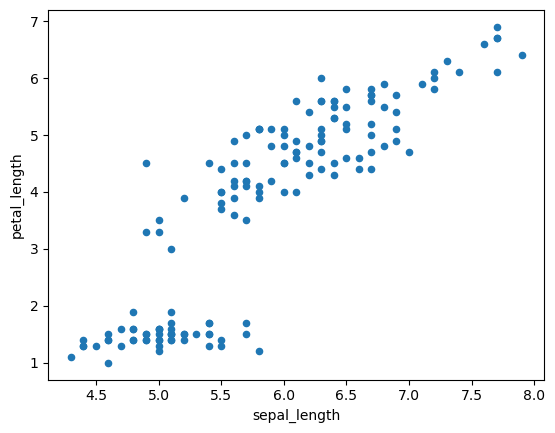
iris.sort_values(list(iris.columns)).plot(style='o', figsize=(10,10));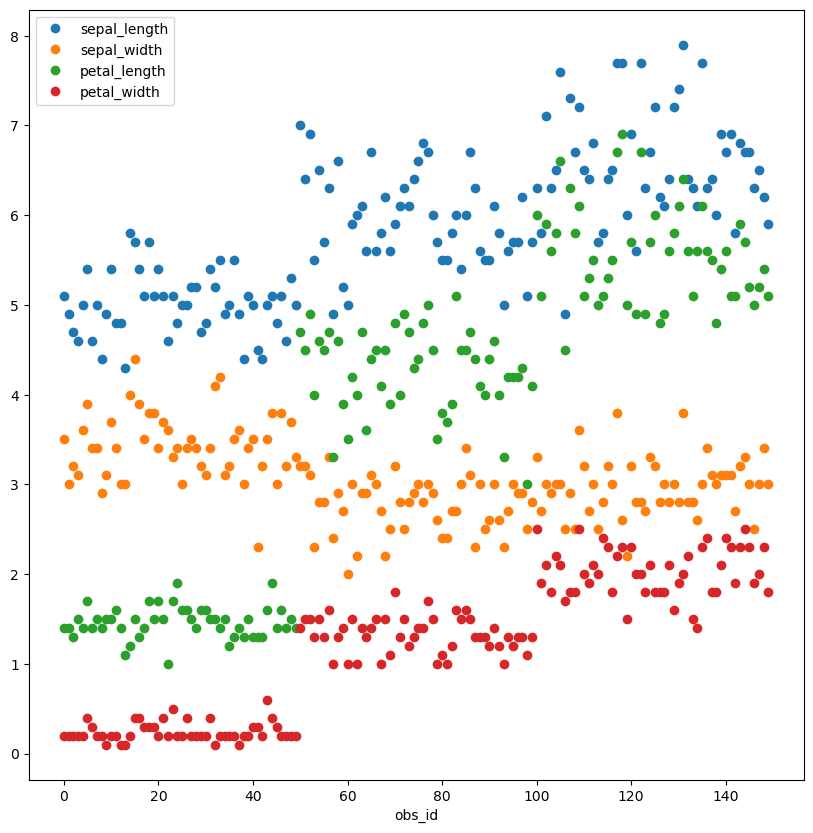
from pandas.plotting import scatter_matrixscatter_matrix(iris, figsize=(10,10));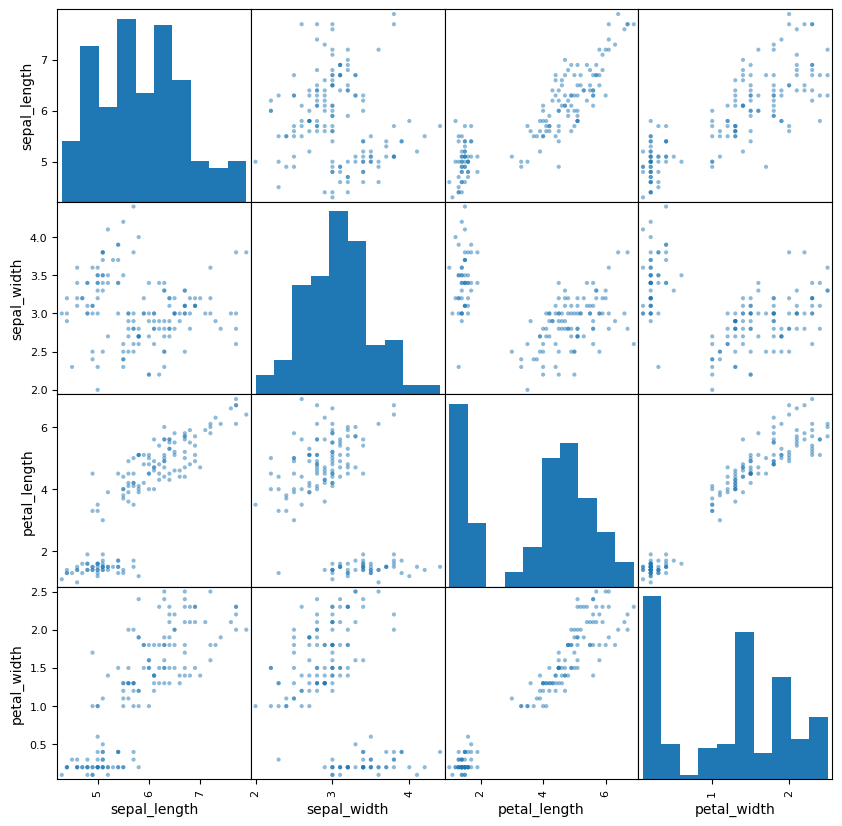
Save to CSV File
Common to save df to a csv file. The full path (path + filename) is required.
There are also options to save to a database and to other file formats,
Common optional parameters: - sep - delimiter - index - saving index column or not
iris.to_csv('./iris_data.csv')Read from CSV File
read_csv() reads from csv into DataFrame
takes full filepath
iris_loaded = pd.read_csv('./iris_data.csv').set_index('obs_id')iris_loaded.head(2)| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| obs_id | |||||
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |